DRDNetPro is a bio-protocol for recovering disease risk-associated pseudo-dynamic networks (DRDNet) from steady-state data. It incorporates risk prediction model of having certain disease, a varying coefficient model, multiple ordinary differential equations, and group lasso estimation to learn a series of networks. This tutorial will provide detailed information for the first step of implementing the DRDNet. The data we will use in this tutorial is named pheno_did.
To begin with, we need to install the following packages in R
Installation
if (!require("devtools")) {
install.packages("devtools")
}
devtools::install_github("chencxxy28/DRDNetPro")## Warning: package 'xgboost' was built under R version 4.1.2Prepare the data for training the agent
The agent in the following tutorial is related to the risk of having hypertension. Before predicting the agent, the users need to prepare the data for training the model. The data is required to be a matrix, which includes the outcome in the first column and all potential predictors in the remaining columns. Do not include the intercept as one column. The outcome in this protocol is in the binary scale (e.g., 0=no disease and 1=disease), which in practice indicates the disease status. All values in the matrix should be numeric. The missing data is allowed in this training process, though it is always recommended to have a complete data as an input. The following working data is from one sample of data in GTEx project and de-identified.
#read the data
pheno_raw<-read.csv("https://github.com/chencxxy28/DRDNetPro/raw/main/vignettes/data/pheno_did.csv")
smoke<-pheno_raw$smoke
pheno_raw<-pheno_raw[,-which(colnames(pheno_raw)=="smoke")]
#rearrange the data
pheno_used<-data.frame(disease=pheno_raw$disease,pheno_raw[,!(colnames(pheno_raw) %in% c("disease","ID"))])
head(pheno_used)## disease V1 V2 V3 V4 V5 V6 V7 V8 V9 V11 V12
## 1 1 1 66 66 32.12 0 0 1 0 1 0 0
## 2 0 0 57 70 33.57 0 0 1 0 0 0 0
## 3 0 0 61 73 25.06 0 0 1 0 0 0 0
## 4 0 0 63 69 29.53 0 0 0 1 0 0 1
## 5 1 0 62 72 30.78 0 0 1 0 1 0 1
## 6 1 1 64 66 32.76 0 0 1 0 0 0 0Build up the training model
There is no unique way to predict the agent (risk of having hypertension). Statistical regression models or machine learning algorithms can be applied. In our program, we allow the users to specify their own method, which includes conventional logistic regression, random forest, and XGBoost. The final imputed agent is supported between 0 and 1, which quantifies the disease risk of interest.
logistic regression
fit<-glm(disease~.,data=pheno_used,family = binomial(link = "logit"))
rate<-fitted(fit)
summary(rate)## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.0563 0.3559 0.5572 0.5587 0.7971 0.9891Random forest
Random forest is an ensemble of decision trees. It builds and
combines multiple decision trees to get more accurate predictions. It’s
a non-linear classification algorithm. Each decision tree model is used
when employed on its own. An error estimate of cases is made that is not
used when constructing the tree. This is called an out of bag error
estimate mentioned as a percentage. There are several R packages
available: one is randomForest, and one is
ranger. Please refer to random
forest for more details. This section provides a demo code based on
the package ranger.
Create the data for ranger, the outcome should be a
factor.
mydata<-pheno_used
mydata$disease<-as.factor(mydata$disease)Run the following code to train the model. Note that some parameters, such as the number of variables, can be further tuned. Please refer random forest for more information.
set.seed(12345)
rf<-ranger(disease~., num.trees=1000,data=mydata,replace=T,write.forest = T,probability = T)
rate.rf<-predict(rf, mydata)$predictions[,2]
summary(rate.rf)## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.02182 0.34699 0.57989 0.55670 0.81908 0.98263XGBoost
XGBoost an efficient implementation of gradient boosting framework.
It provides built-in k-fold cross-validation Stochastic GBM with column
and row sampling (per split and per tree) for better generalization,
includes efficient linear model solver and tree learning algorithms, and
supports various objective functions, including regression,
classification and ranking. Please refer to gradient boosting for
more details. This section provides a demo code based on the package
XGBoost.
XGBoost requires a specific type of input data, which can be run by the following code
mydata<-pheno_used
# variable names
features <- setdiff(names(mydata), "disease")
# Create the treatment plan from the training data
treatplan <- vtreat::designTreatmentsZ(mydata, features, verbose = FALSE)
# Get the "clean" variable names from the scoreFrame
new_vars <- treatplan %>%
magrittr::use_series(scoreFrame) %>%
dplyr::filter(code %in% c("clean", "lev")) %>%
magrittr::use_series(varName)
# Prepare the training data
features_train <- vtreat::prepare(treatplan, mydata, varRestriction = new_vars) %>% as.matrix()
response_train <- mydata$diseaseTune the number of trees that minimizes the error
set.seed(12345)
xgb.fit1 <- xgb.cv(
data = features_train,
label = response_train,
nrounds = 1000,
nfold = 5,
objective = "binary:logistic", # for regression models
verbose = 0 , # silent,
early_stopping_rounds = 10 # stop if no improvement for 10 consecutive trees
)
vect=xgb.fit1$evaluation_log %>%
dplyr::summarise(
ntrees.train = which(train_logloss_mean == min(train_logloss_mean))[1],
rmse.train = min(train_logloss_mean),
ntrees.test = which(test_logloss_mean == min(test_logloss_mean))[1],
rmse.test = min(test_logloss_mean),
)Run the final trained model to obtain the predicted agent
set.seed(12345)
#final estimate
xgb.fit.final <- xgboost(
data = features_train,
label = response_train,
nrounds = as.numeric(vect[3]),
objective = "binary:logistic", # for regression models
verbose = 0 # silent,
)
rate.xgb = predict(xgb.fit.final, features_train,type="response")
summary(rate.xgb)## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.0809 0.3987 0.5647 0.5490 0.7681 0.9278Predict and evaluate the agent
When the agent is imputed, it is better to check its prediction performance. Note that the pseudo-dynamic network is sensitive to the values of agent, and the agent with good prediction to the disease risk is more preferred. To check its performance, we can use ROC and AUC value. For evaluation purpose, we need to create a training data and testing data. The training data is used for data fitting, whereas the testing data is used for prediction evaluation. Below, we consider 90% of data for training and 10% of data for testing.
set.seed(12345)
sample.ind<-sample(1:nrow(pheno_used),round(0.9*nrow(pheno_used))) #id used for training
pheno_used_training<-pheno_used[sample.ind,]
pheno_used_testing<-pheno_used[!(1:nrow(pheno_used) %in% sample.ind),]
fit_train<-glm(disease~.,data=pheno_used_training,family = binomial(link = "logit"))
test_prob = predict(fit_train, newdata = pheno_used_testing, type = "response")
test_roc_specific =roc(pheno_used_testing$disease ~ test_prob, plot = TRUE, print.auc = TRUE)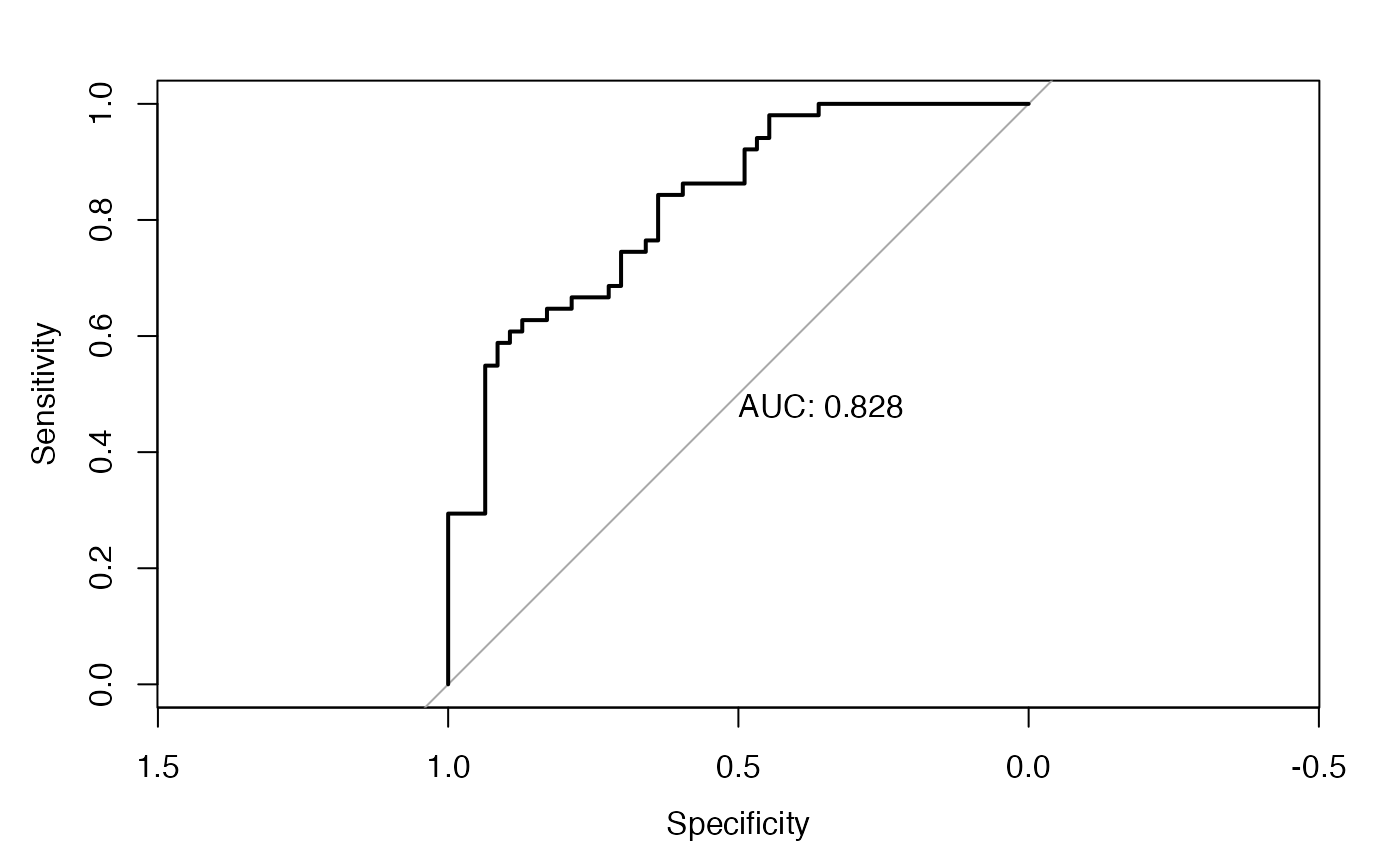
The AUC value is around 0.83, which is desired. Similar procedure could be applied to the case with random forest or XGBoost. After the agent imputation, we can move forward to the stage of network learning, which is illustrated in Tutorial 2. Before the end of this tutorial, let us create a phenotypical data consisting of the imputed agent for the use in next tutorial.
pheno_subject_id<-substring(pheno_raw$ID,6,10)
pheno_together<-data.frame(cbind(id=pheno_subject_id,rate,smoke,pheno_raw$disease))
colnames(pheno_together)<-c("id","rate","smoke","hyper")
head(pheno_together)## id rate smoke hyper
## 1 1117F 0.858620894429252 1 1
## 2 111CU 0.59473507595632 1 0
## 3 111FC 0.528193073054949 0 0
## 4 111VG 0.890841096353929 0 0
## 5 111YS 0.946066339003859 1 1
## 6 1122O 0.694087589148978 1 1
write.csv(pheno_together,"pheno_together.csv",row.names =F)This is the end of the Tutorial 1, please visit Tutorial 2 for the next step of network learning